Clustering of PCA results using TRENDanalysis
Parameters
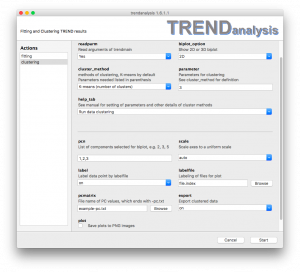
- The
clusteringmenu of TRENDanalysis is used to visualize and cluster PCA results calculated by by Trendmain or TREND NMR - When the
readparmoption is turned on (by choosingYes),TRENDanalysisreads PC values calculated byTrendmainorTREND NMR. Ifreadparmis turned off (by choosingNo), the data will be read instead from the file specified in thepcmatrixfield. biplot_optionindicates which PCA biplots to display. Choices are2D,3D,Both 2D and 3D, andNone.cluster_methodprovides options of clustering algorithms, The default isK-means. Parameters needed are listed in parentheses. See details under Clustering methodsparametersets the parameter for clustering. Seecluster_methodandhelp_tabfor details.help_tab
By defaulthelp_tabis set asRun data clusteringso that TRENDanalysis will run display one or two biplots onceStartis pressed. . Other options trigger listing of the parameters relevant to thecluster_methodselected. Pressingstartwill launch the help file in html format.pcn
In this field, please list your choice of components to appear in the biplot, most often 1-3. But could be another combintaion, such as 2,3,5. At least 3 components must be selected. If 4 or more are selected, only the first 3 components will be plotted in 2D or 3D biplots. However, clustering results will be calculated using 3 or more components selected.
The syntax is equivalent to specifying pages in a print dialog. For example,1, 3-5, 7means selectinig components 1, 3, 4, 5, 7.scalecontrols scaling of output biplots. Ifuniformis chosen then in 2D or 3D biplots the axes will use the same scale, otherwise each axis will be scaled automatically.label
whenlabelturns the labeling of data points in biplots on or off.labelfilerefers to the file containg a list of labels for the points. The format of this file is identical tofile.index(See the manual of trendmain for the format offileindex.) Note the sequence in a label file must be identical to its corresponding [fileindex] file. When no file is chosen forlabelfile, TRENDanalysis will use the file names to label the data points in biplots.pcmatrixWhenreadparmis turned off,pcmatrixis used to read PCA results calculated by TRENDmain or TREND NMR. The file name ends in-pc.txt.exportset to on causes clustering results to be saved asprefix-cluster_2d.txtorprefix-cluster_3d.txt- plot
When this option is checked, 2D and/or 3D biplots will be saved as images in the.pngfile format.
Choice of clustering method:
- K-means (number of clusters)
The K-means algorithm requires the user to specify the number of clusters in parameter option. By default it is set as 3. See K-means for details.
- Agglomerative (number of clusters)
Similar to K-means, agglomerative clustering also needs the user to specify the number of clusters. See Agglomerative Clustering for details.
- Affinity Propagation (preference)
Affinity Propagation does not require prior-knowledge of number of clusters. Instead, preference is used to choose exemplars. Choose None in the parameter filed sets preference to the median of input similarities. See affinity_propagation for details.
- DBSCAN (min_sample)
This option performs Density-Based Spatial Clustering of Applications (DBSCAN) clustering. The min_sample parameter defines the number of samples to be considered as a core point. See DBSCAN for details.
- Mean Shift (bandwidth)
This option applys mean shift clustering. The parameter can be set to None. See Mean Shift for details.
